Supplying radial coupling terms of the PES
- Hubert Jóźwiak
The reading process is controlled by both the input file and the radial_coupling_terms_mod module.
To avoid recompilation of the code whenever the radial coupling terms file is switched, we use a fixed format for radial coupling terms file. Below, we present a screenshot of the oH2-He-radialterms.dat file which can be found in the oH2-He-radialterms.zip archive in the ref directory:
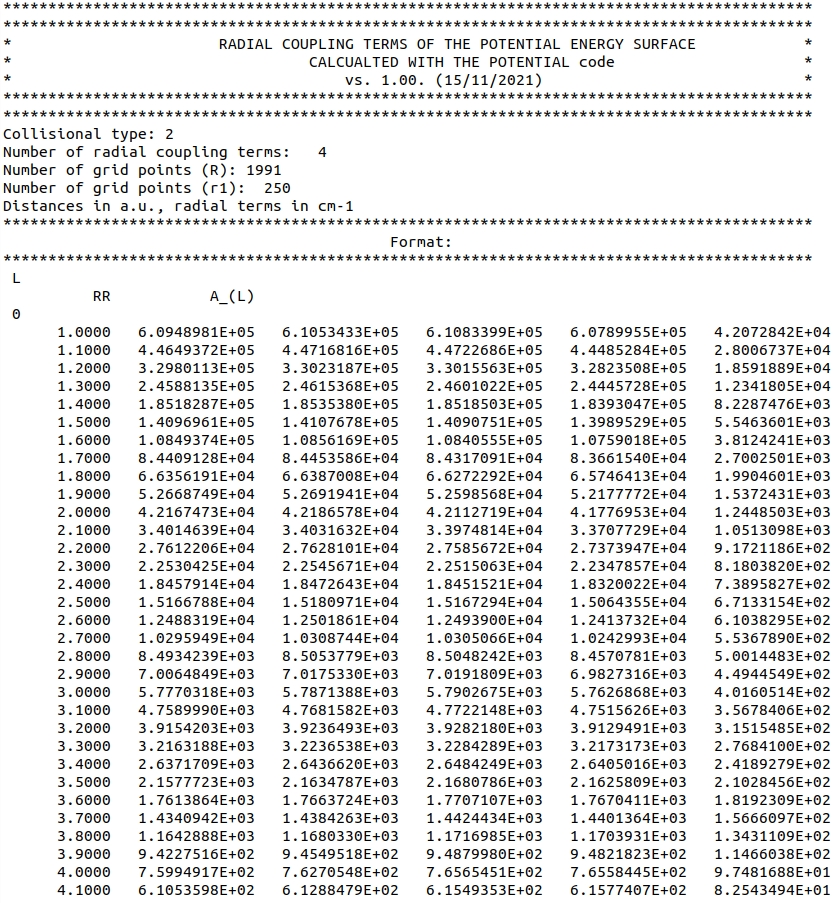
The file starts with n_skip_lines = 17 lines which describe the file and are ignored by the code. The reading procedure is continued using two loops: the first one over number_of_legendre_indices (which matches the size of legendre_indices), and the second one over number_of_r_points (the number of points on the \( R \)-grid).
The code assumes that each loop over number_of_legendre_indices begins with a current value of} \( \lambda \), and that the radial coupling terms are ordered in columns, where the first column corresponds to the \( R \)-grid. Additional loop over total_number_of_coupling_terms reads radial terms which couple different \( v, j \) and \( v', j' \) states.
- \(v=0,\,j=1,\,v'=0,\,j'=1\),
- \(v=0,\,j=1,\,v'=0,\,j'=3\),
- \(v=0,\,j=1,\,v'=0,\,j'=5\),
- \(v=0,\,j=1,\,v'=0,\,j'=7\),
- \(v=0,\,j=1,\,v'=1,\,j'=1\),
The order of columns in the radial terms file corresponds to the values supplied in the vib_couplings, rot_couplings, vib_prime_couplings, rot_prime_couplings arrays in the input file.
Reducing the number of radial coupling terms
The user can supply a larger set of terms that describe the coupling between different rovibrational states (\( v, j \) and \( v', j' \) ) than needed.
This is exactly the case here, as some of the terms in this file describe coupling off-diagonal with \(v\), although the corresponding input file
involves only three rotational levels from the \(v=0\) manifold.
If this is the case, the reduce_radial_coupling_terms subroutine from radial_coupling_terms_mod module picks only the necessary terms, i.e. terms that describe couplings between all levels in the rovibrational basis set.
Note that the procedure will be skipped if total_number_of_coupling_terms \( <= \) number_of_basis_levels \( \cdot \) (number_of_basis_levels+1)\(/2\).